lv sgrna | In vivo CRISPR screens reveal Serpinb9 and Adam2 as lv sgrna The human CRISPR Brunello lentiviral pooled libraries were designed using optimized metrics which combine improved on-target activity predictions (Rule Set 2), with an off-target score, the Cutting Frequency Determination (CFD).
Left ventricular non-compaction (LVNC) is a rare cardiomyopathy in which the two-layered myocardium has an abnormally thick sponge-like, noncompacted trabecular layer and a thinner, compacted myocardial layer. It is characterized by prominent trabeculae, showing continuity between the deep trabecular recesses and the .
0 · In vivo CRISPR screens reveal Serpinb9 and Adam2 as
1 · Human CRISPR Knockout Pooled Library (Brunello)
Left ventricular ejection fraction (LVEF) is the central measure of left ventricular systolic function. LVEF is the fraction of chamber volume ejected in systole (stroke volume) in relation to the volume of the blood in the ventricle at the end of diastole (end-diastolic volume).
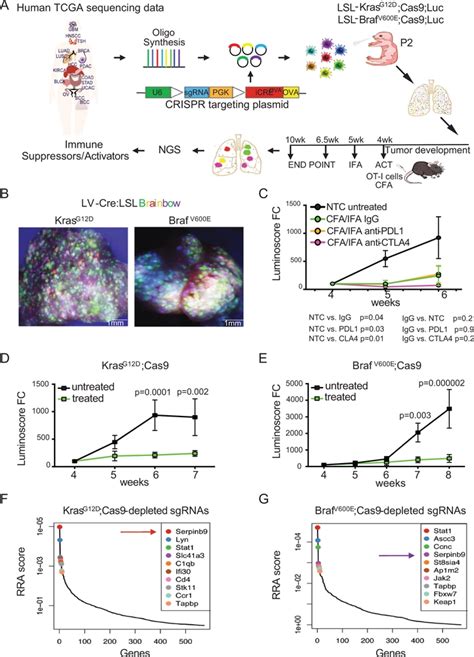
A Experimental workflow of the in vivo CRISPR screen to identify immune-modulatory cancer genes. A lentiviral sgRNA library targeting mouse homologs of 573 human .We would like to show you a description here but the site won’t allow us.We would like to show you a description here but the site won’t allow us.The human CRISPR Brunello lentiviral pooled libraries were designed using optimized metrics which combine improved on-target activity predictions (Rule Set 2), with an off-target score, .
A Experimental workflow of the in vivo CRISPR screen to identify immune-modulatory cancer genes. A lentiviral sgRNA library targeting mouse homologs of 573 human genes, which are associated with.The human CRISPR Brunello lentiviral pooled libraries were designed using optimized metrics which combine improved on-target activity predictions (Rule Set 2), with an off-target score, the Cutting Frequency Determination (CFD). Genomic integration of LV-delivered sgRNA constructs for use as sequencing barcodes can serve as a surrogate for identifying causal genes: Off-target effects related to Cas9 toxicity and promiscuous Cas9/sgRNA binding [27,28] Gene targeting efficiency varies owing to sgRNA on-target efficacy and chromatin state Cells were infected with pools of four lv-sgRNA or individual shRNAs. A z-score cutoff for a p-value < 0.01 was used to define a positive hit, with all scoring genes indicated within the box.
Based on these observations, here we developed hybrid particles for Cas9 mRNA and sgRNA co-delivery with normal capsid assembly efficiency. We further improved LVs for integrated gene expression by including an aptamer sequence in lentiviral genomic RNA, which improved lentiviral particle production and enhanced LV genomic RNA packaging. Simultaneous transfection of sgRNA-expressing lentiviral transfer plasmid DNA into the VLP-producing cells produced hybrid VLP/LV particles containing SpCas9 mRNA and sgRNA-expressing LV genomic RNA. Since the integrase bears a D64V mutation, the sgRNA-expression cassette will not be integrated.
In vivo CRISPR screens reveal Serpinb9 and Adam2 as
To verify the effect of KIBRA in MPC5 cells, LV-Cas9 was seeded in MPC5 cells with an MOI of 20 after 5–7 days of puromycin selection at a final concentration of 0.6 μg/ml; LV-sgRNA was then . To investigate this hypothesis, we utilized LV transduction of human HSPCs as a lower-stress manipulation and evaluated the impact of G-CSF treatment on these cells. . The sequences around the sgRNA target site of AAVS1, ITGB2, CXCR4 and G-CSFR genomic regions were uploaded to Microhomology-Predictor of CRISPR RGEN tools . The latter approach for sgRNA packaging relies on the high affinity between Cas9 and its sgRNA, which enables gag–Cas9 fusions to be loaded with sgRNAs prior to packaging within VLPs. These Cas9-VLPs enabled up to 90% editing in Jurkat cells and up to 70% editing in primary human T cells. With the emergence of CRISPR-Cas9 gene editing technology, functional genetic single-guide RNA (sgRNA) libraries now exist that are in theory capable of triggering biallelic insertion-deletion (indel) mutations in most genes in the human genome (Shalem et al., 2014, Wang et al., 2014).
A Experimental workflow of the in vivo CRISPR screen to identify immune-modulatory cancer genes. A lentiviral sgRNA library targeting mouse homologs of 573 human genes, which are associated with.The human CRISPR Brunello lentiviral pooled libraries were designed using optimized metrics which combine improved on-target activity predictions (Rule Set 2), with an off-target score, the Cutting Frequency Determination (CFD).
Genomic integration of LV-delivered sgRNA constructs for use as sequencing barcodes can serve as a surrogate for identifying causal genes: Off-target effects related to Cas9 toxicity and promiscuous Cas9/sgRNA binding [27,28] Gene targeting efficiency varies owing to sgRNA on-target efficacy and chromatin state Cells were infected with pools of four lv-sgRNA or individual shRNAs. A z-score cutoff for a p-value < 0.01 was used to define a positive hit, with all scoring genes indicated within the box. Based on these observations, here we developed hybrid particles for Cas9 mRNA and sgRNA co-delivery with normal capsid assembly efficiency. We further improved LVs for integrated gene expression by including an aptamer sequence in lentiviral genomic RNA, which improved lentiviral particle production and enhanced LV genomic RNA packaging.
Simultaneous transfection of sgRNA-expressing lentiviral transfer plasmid DNA into the VLP-producing cells produced hybrid VLP/LV particles containing SpCas9 mRNA and sgRNA-expressing LV genomic RNA. Since the integrase bears a D64V mutation, the sgRNA-expression cassette will not be integrated. To verify the effect of KIBRA in MPC5 cells, LV-Cas9 was seeded in MPC5 cells with an MOI of 20 after 5–7 days of puromycin selection at a final concentration of 0.6 μg/ml; LV-sgRNA was then . To investigate this hypothesis, we utilized LV transduction of human HSPCs as a lower-stress manipulation and evaluated the impact of G-CSF treatment on these cells. . The sequences around the sgRNA target site of AAVS1, ITGB2, CXCR4 and G-CSFR genomic regions were uploaded to Microhomology-Predictor of CRISPR RGEN tools .
The latter approach for sgRNA packaging relies on the high affinity between Cas9 and its sgRNA, which enables gag–Cas9 fusions to be loaded with sgRNAs prior to packaging within VLPs. These Cas9-VLPs enabled up to 90% editing in Jurkat cells and up to 70% editing in primary human T cells.
Human CRISPR Knockout Pooled Library (Brunello)
 .jpg)
Compare Car Insurance online and see how LV= measures up to other insurance companies. Compare features and find out more about our cover today.
lv sgrna|In vivo CRISPR screens reveal Serpinb9 and Adam2 as